Large foundation models have demonstrated remarkable potential in biomedical applications, offering promising results on various benchmarks and enabling rapid adaptation to downstream tasks with minimal labeled data requirements. However, significant challenges persist in implementing these models in clinical settings. Even advanced models like GPT-4V show considerable performance gaps in multimodal biomedical applications. Moreover, practical barriers such as limited accessibility, high operational costs, and the complexity of manual evaluation processes create substantial obstacles for clinicians attempting to utilize these state-of-the-art models with private patient data.
Recent developments in multimodal generative AI have expanded biomedical applications to handle text and images simultaneously, showing promise in tasks like visual question answering and radiology report generation. However, these models pose challenges in their clinical implementation. Large models’ resource requirements pose deployment challenges in computational costs and environmental impact. Small Multimodal Models (SMMs), while more efficient, still show significant performance gaps compared to larger counterparts. Additionally, the lack of accessible open-source models and reliable evaluation methods for factual correctness, particularly concerning hallucination detection, creates substantial barriers to clinical adoption.
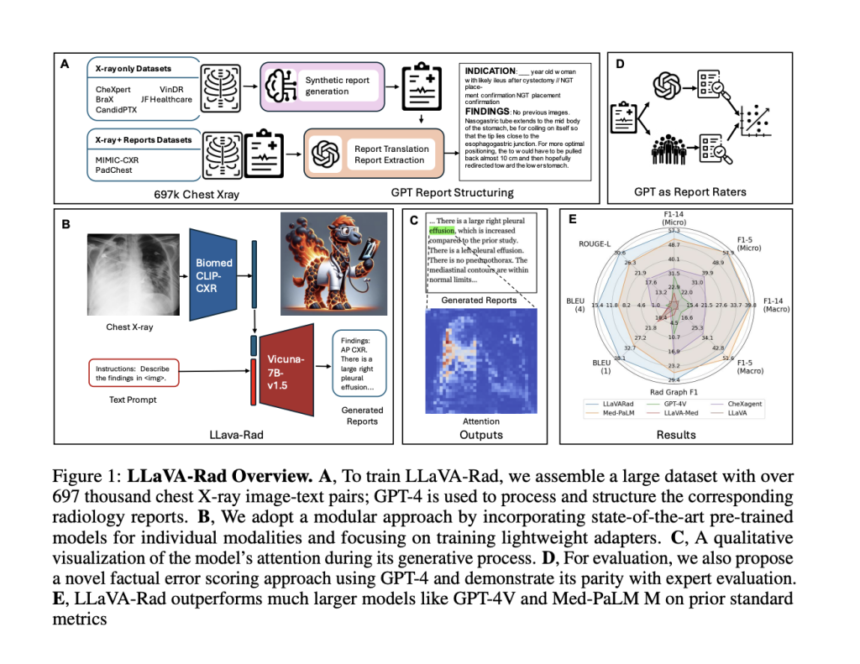
Researchers from Microsoft Research, the University of Washington, Stanford University, the University of Southern California, the University of California Davis, and the University of California San Francisco have proposed LLaVA-Rad, a novel Small Multimodal Model (SMM), alongside CheXprompt, an automatic scoring metric for factual correctness. The system focuses on chest X-ray (CXR) imaging, the most common medical imaging examination for automatically generating high-quality radiology reports. LLaVA-Rad is trained on a dataset of 697,435 radiology image-report pairs from seven diverse sources, utilizing GPT-4 for report synthesis when only structured labels were available. The system demonstrates efficient performance, requiring just a single V100 GPU for inference and completing training in one day using an 8-A100 cluster.
LLaVA-Rad’s architecture represents a novel approach to Small Multimodal Models (SMMs), achieving superior performance despite being significantly smaller than models like Med-PaLM M. The model’s design philosophy centers on decomposing the training process into distinct phases: unimodal pretraining and lightweight cross-modal learning. The architecture utilizes an efficient adapter mechanism to ground non-text modalities into the text embedding space. The training process unfolds in three stages: pre-training, alignment, and fine-tuning. This modular approach uses a diverse dataset of 697,000 de-identified chest X-ray images and associated radiology reports from 258,639 patients across seven different datasets, enabling robust unimodal model development and effective cross-modal adaptation.
LLaVA-Rad shows exceptional performance compared to similar-sized models (7B parameters) like LLaVA-Med, CheXagent, and MAIRA-1. Despite being substantially smaller, it outperforms the leading model Med-PaLM M in critical metrics, achieving a 12.1% improvement in ROUGE-L and 10.1% in F1-RadGraph for radiology text evaluation. The model maintains consistent superior performance across multiple datasets, including CheXpert and Open-I, even when tested on previously unseen data. This performance is attributed to LLaVA-Rad’s modular design and data-efficient architecture. While Med-PaLM M shows marginally better results (<1% improvement) in F1-5 CheXbert metrics, LLaVA-Rad’s overall performance and computational efficiency make it more practical for real-world applications.
In this paper, researchers introduced LLaVA-Rad which represents a significant advancement in making foundation models practical for clinical settings, offering an open-source, lightweight solution that achieves state-of-the-art performance in radiology report generation. The model’s success stems from its comprehensive training on 697,000 chest X-ray images with associated reports, utilizing GPT-4 for dataset processing and implementing a novel three-stage curriculum training method. Moreover, the introduction of CheXprompt solves the crucial challenge of automatic evaluation, providing accuracy assessment comparable to expert radiologists. These developments mark a significant step toward bridging the gap between technological capabilities and clinical needs.
Check out the Paper and GitHub Page. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter and join our Telegram Channel and LinkedIn Group. Don’t Forget to join our 75k+ ML SubReddit.
The post Microsoft AI Researchers Release LLaVA-Rad: A Lightweight Open-Source Foundation Model for Advanced Clinical Radiology Report Generation appeared first on MarkTechPost.


 Recommended Open-Source AI Platform: ‘IntellAgent is a An Open-Source Multi-Agent Framework to Evaluate Complex Conversational AI System’ (Promoted)
Recommended Open-Source AI Platform: ‘IntellAgent is a An Open-Source Multi-Agent Framework to Evaluate Complex Conversational AI System’ (Promoted)